Horos Horos - a free, open medical image viewer. The goal of the Horos Project is to develop a fully functional, 64-bit DICOM viewer for OS X based on OsiriX Similar to OsiriX MD and for free. Free Hiarcs Chess Explorer (mac Version) Programs Skype Add In For Outlook 2011 For Mac Omnisphere Mac Osx Keygen Utorrent V1.8.9 Pre-cracked For Mac Deus Ex Mankind Divided For Mac Activate Dicom Editing Osirix Lite For Mac Widsmob Viewer 2.4.1240 For Mac Best 4k Display For Mac Pro 2013 For Mac Google Chrome. Activate dicom editing osirix lite for mac. OsiriX Lite is at the same time a DICOM PACS workstation for medical imaging and an image processing software for medical research (radiology and nuclear imaging), functional imaging, 3D imaging, confocal microscopy and molecular imaging. OsiriX Lite is an advanced DICOM viewer application for MacOS X. It is compatible with DICOM files and PACS networks. OsiriX is able to receive DICOM images (STORE SCP - Service Class Provider) from a PACS network. OsiriX has been specifically designed for image fusion, cardiac CT, multi-series MRI exams and 3D reconstructions. MAC OSIRIX if the best free dicom viewer available (also one of the best in general among free and not free). Using tis software you can export dicom to jpeg and also do video, 3D reformats, MPR.
Mar 22, 2013
Configuring And Operating An OsiriX Teaching File Server
Updated in December 2015
In this guidance document I'll describe how to configure and operate an OsiriX-based Teaching File Server that will enable you to share your cases with colleagues.
If you and your colleagues put your individual servers on an Internet-based network , each network member will, in effect, have a Teaching File consisting of the cases on all the servers.
How this works will become apparent as you read this document.
Contents
- Hardware and Software Considerations
- Network Considerations
- Backing up the data on your Server
- Getting and anonymizing DICOM cases
- Configuring OsiriX on your Server
- Understanding a JPEG 2000 Teaching Server Network
- Querying a Server for Teaching Cases
- Exporting a case to share with colleagues
Hardware and Software Considerations
A Mac mini or iMac is fine as a Server. There is no need to utilize OS X Server. To use OsiriX for the purpose described in this document, you'll need to purchase the non-free version. OsiriX Lite, the free version, has too many functional limitations, substantially increased since I wrote the first version of this guidance. Explore the Pixmeo Website for purchase options, including the possibility of a Site License for your department or practice.
If you can afford it, purchase a device with a SSD drive, and at least 8GB of RAM. The combination of a SSD drive and El Capitan (the latest version of the OS X Operating System as of Late 2015) will optimize the serving of images.
The average size of a Teaching Case on my Server is around 30-50 MB, the case being stored in a compressed state (JPEG 2000 compression). Many of my cases contain multiple CT, CT-PET and/or MR Series, each Series sometimes consisting of several hundred thin-section images.
If you choose a Mac mini, you can operate it via remote control (if necessary) using the Screen Sharing VNC application that comes with OS X. Other third-party VNC applications are available for this purpose but are not, as far as I know, better than Screen Sharing for this purpose.
Note that you can use iCloud and Back to my Mac for screen sharing as long as both devices are configured to use the same iCloud account. A guide for this is here. However, in my experience, this is not at all a reliable or efficient method.
I suggest the following password-protected configuration (System Preferences) for Screen Sharing:
Here is an image of me working with my (non-local) Mac mini Server from home:
For an OsiriX server, you need to institute a number of actions and specify particular system-wide preferences.
General instructions and guidance are available at this Web page.
Here is a summary of the things you should do:
I want to be sure that OsiriX is always running and is restarted in the event of an elecricity failure or an unexpected application crash.
I use the application Lingon for this purpose. Here is a sceenshot of it running on my server:
If you use Lingon for this purpose, specify that OsiriX is initiated at login/load and is 'kept running.'
In System Preferences: Energy Saver, select ' Start up automatically after a power failure.' Move the slider for 'Computer Sleep' to 'Never' and select 'Wake for network access.' Do not select ' Put hard disks to sleep when possible.'
In System Preferences: Users&Groups: Login items on your Mac, make sure that OsiriX is in the List. If you use Lingon, then it need not be.
If your Server is a Mac mini which is not connected to a display, go to System Preferences: Desktop&Screen Saver, and change the option on the Screen Saver to
Neverwhich otherwise sometimes interferes with Screen Sharing. Change your Destop Background to a simple solid color.In System Preferences, select automatic log-in (Users & Groups, Login Options, Automatic login)
Activate the 'Server mode' in OsiriX. This mode will hide the GUI, and OsiriX will never present a blocking window to the user (nobody will look at the server monitor to click the 'OK' button…). Go to OsiriX Preferences: Listener, Server mode.
When an application on the Mac crashes, a Crash Reporter often comes up, asking you to respond and, optionally, to send a report to Apple. For a Server, you're not there to respond to this. To prevent this from happening, open the Terminal utility, select-and-copy the following, paste it at the command line and hit 'return':
defaults write com.apple.CrashReporter DialogType Server
Like this:
You can still see and inspect the Crash Reports in the Console app if you wish.
If you ever want to change this back to the default mode, type this at the command line:
defaults write com.apple.CrashReporter DialogType Developer
Networking Considerations
The Server needs to have a static (fixed) IP address and the fastest connection speed to the Internet as reasonably achievable. There are three basic options for accomplishing this:
- Locating your Server at home
- Locating your Server at work
- Locating your server at a Colocation Facility
Home server
One purchases a package from an Internet Service Provider that includes a Static IP address(es) and a certain bandwidth. Availability and affordability are considerations for this option.
Server at work
In this scenario, you server will be physically connected by ethernet cable to the wired hospital Local Area Network (LAN). Your IT department should be able to reserve a particular IP address for your server by means of so-called dynamically assigned static IP addresssing. Ask for this. This is very convenient as you'll then easily be able to send cases on your laptop to it from anywhere. My Mac mini Server is in my office and I DICOM-send cases to it from anywhere in the hospital such as the Reading Room where I work each day. If your Server's LAN IP address is fixed you'll also be able to start the OsiriX Web Server with that address as the Public address and the Web Portal will then always be available at that address, which you can then share with departmental colleagues.
Your hospital may also provide a means to connect to the hospital network from the outside via a VPN application of some kind . If so, you'll first connect to the network via the VPN and then have access to your Server via its fixed LAN address. The following image shows a browser-based VPN application I use to connect to my institution's network:
Connecting to your server
In order to connect to your office Server, you can create a Virtual Private Network using an application called ZeroTier. Review the guidance document on this website about this.
In this scenario, your Server will be assigned a network-associated static IP address and other clients in the private network only will be able to connect to it.
Your Server's OsiriX instance may have multiple IP addresses simultaneously, one of which is your address on the ZeroTier One network, as shown in this portion of my Preferences:Listener panel:
The advantages of this option are:
- You will have access to your server at the hospital on a high-speed and high-bandwidth wired LAN
- You'll have your institution's fast bandwidth connection to the Internet to facilitate retrieval of cases from the Server from outside the hospital.
- You'll be able to connect to your Server from anywhere (including outside the hospital) via Screen Sharing using the ZeroTier One network IP address
- You'll be able to connect to the Server's OsiriX Web Portal via either the Portal's LAN IP address (while at the hospital) or via the ZeroTier One network (while outside the hospital)
- You'll not have to purchase a static IP address package from your Internet Service Provider
Server at a Colocation Facility
At first, this idea may seem silly as your computer has to be shipped to another physical location. However, there are entities that specialize in colocating Mac minis, such as MacStadium and Mac mini Vault. They provide you with a static IP address, and offer fast Internet connections at a reasonable price. You can manage your server remotely using Screen Sharing (or a VPN application they'll provide you) as I mentioned above.
Update: May 2014
I have moved my Mac mini from the hospital to a colocation facility— MacStadium. The upload speed from there to the Internet is much faster than with the usual home setup, which is great for serving up cases. Here is a screenshot showing upload and download speeds at the facility. (You can test your own speeds at: beta.speedtest.net)
Backing up the Server
I strongly suggest that you employ two concurrent means of backing up your OsiriX Data Folder: 1) to an attached external disk utilizing software such as Time Machine and 2) to an account in the Cloud. Options for cloud storage are Crash Plan, Backblaze, and an online cloud-storage service using the application Arq. It's easy to schedule automatic backups (once a day) with these.
Getting and anonymizing DICOM cases
If your PACS application permits you to export a case in DICOM format with one click, the most efficient method is to ask your IT department to assign your laptop (or another device other than your Server) a static IP address as well, as I described above. Then, register your OsiriX laptop as a DICOM-node on the PACS administration module. When your laptop is connected to the network via an ethernet cable, you will: 1) DICOM-send the case to your laptop OsiriX, 2) anonymize the case on your laptop, and 3) DICOM-send the anonymized case to your Server. Otherwise, you'll have to use another method such as a USB thumb drive to transfer the case to your laptop.
Remember: A non-anonymized case should not be on your Server at any time.
Here is a screenshot of me sending a case to my laptop OsiriX directly from our Philips iSite PACS:
I always have my laptop connected by ethernet cable to the LAN at the Workstation I'm working at, so I can do this as I perform my daily work in the Reading Room. If I have time right then, I'll anonymize it, check the meta-data again to verify that the anonymization of personal information is complete, and DICOM-send it to my Server. Otherwise, I'll do this step another time.
Anonymizing cases – three methods
Using the OsiriX anonymizer
Here is the scheme I currently use. It removes common sources of Personal Information, including that in Private Tags that vendors often employ.
Add to these as necessary as you inspect the Metadata as I indicate below.
Important: When you use the 'Merge' command in OsiriX to merge all the Studies/Series of a particular case before anonymizing it, the component Studies will be split again when you DICOM-send or Retrieve it from the Server. This is annoying! Prevent this by putting an appropriate number in the DICOM Tag 'StudyInstanceUID' as indicated above in the graphic.
This number should be derived from your personal UID root (prefix) that you use for all the cases in your Teaching File(s). Review this guidance for information about getting this prefix and how to use it. It's easy!
The Diagnosis you insert should not be longer than 64 characters to comply with the Dicom Standard.
With all the DICOM tags I've added to this scheme most cases are anonymized such that all personal information is removed. But not always. Scroll through the Metadata of a Series of a case to be sure that none is left. If any remains, use the next manual method to remove it. You can also use the manual method to edit any particular field(s) in the DICOM Header (Metadata).
The Manual Method
OsiriX permits one to edit any particular field in the DICOM header. This is a three-step process as indicated below:
- Activate the editing function by clicking on
edit. - Double-click on an entry to edit it and hit return. Edit other items as necessary.
- Click
Applyspecifying theStudylevel so that all the Series of the Case are changed.
Using Dicom Anonymizer to anonymize cases
If you do not use OsiriX to anonymize a case, a great application to do this is Dicom Anonymizer. Indeed, it's easiest to anonymize a case on your desktop with this before importing it into OsiriX.
Review my guidance document on this web site for details on its use.
Removing personal patient information embedded in an image
Sometimes, for example, with an ultrasound image series, PHI is embedded in the image and cannot be removed by editing the DICOM header. In this case, the relevant portion of the image has to be removed, the resultant image saved as a separate Series, and the original Series deleted.
The process is as follows:
- Define the area to be retained with the
rectangular ROI tool. In the image below, I'm using this technique to exclude extraneous markers from a bedside chest radiographic image. The same process may be used for an ultrasound or nuclear medicine series containing many images. - Click on the Shutter Tool icon in the toolbar. If the icon is not there, right-click on the toolbar, choose
Customize Toolbarand drag it there. - This image demonstrates what you see just before you invoke the Shutter Tool
Next steps…
- Now that everything outside the ROI has been removed, use the Zoom and Pan tools to resize and center the resultant image as necessary in the window.
- Hit
Command-Eto save this as a new Series (named as you choose) in the case. For Image Format, specifyas displayed in 16-bit BWbefore clicking OK. - Delete the original series.
If you want to view an excellent video of this process applied to an ultrasound examination, do so right here (Credit: Dr. Mary Roddie, OsiriX U.K. User Group):
If you have a single jpg' alt='image with embedded personal information that you want to import into an OsiriX case, use the Preview application that comes with every Mac to crop out the information (or blacken it) and then import it into the case with the OsiriX menu item: Plugins...Database...JPEG to DICOM utility, specifying the Meta-data: from the selected study in Database window option when doing so.
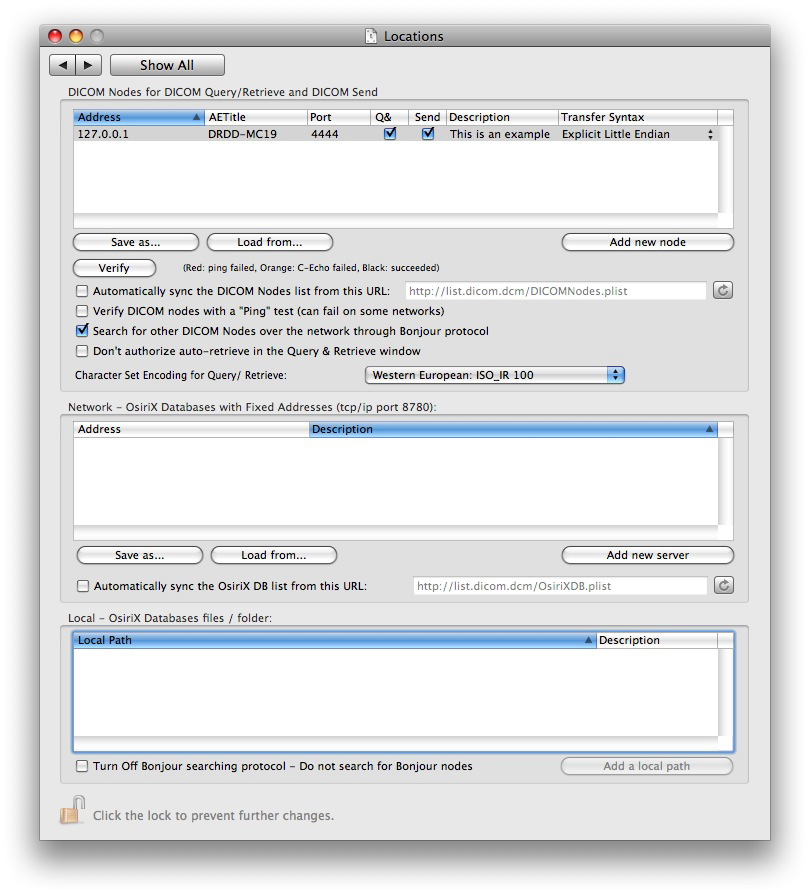
Configuring OsiriX on the server
You need to configure certain items in OsiriX Preferences differently for a Server compared to a Retrieving Workstation. The scenario is analogous to that in your department: Your PACS Server delivers images on demand to Workstations; the former is equivalent to your OsiriX Teaching File Server and the latter are the devices retrieving images from it. The options you should specify in your OsiriX Preferences are indicated on the following screenshots from my Server:
Do not choose the option to broadcast the existence of your Server via the Bonjour protocol. If you do, any person with OsiriX on the network may see your Server's Dicom-node parameters (AE Title, IP address and Port) by hovering the cursor over it in the Sources List and try to query your Server for cases via the Query/Retrieval window, as explained further below:
Instead of this, you could manually add it to the List of Shared Databases on your laptop's OsiriX as shown here:
When you specify 'WADO' above, you'll need to specify additional items as indicated here:
It's very important to specify retrieval using the **Original Syntax** which is JPEG 2000. If you do not, the server will decompress the images before sending them diminishing the transfer efficiency. In addition, the decompressed images will then remain on the server in a decompressed state, slowing the retrieval speed for subsequent retrieval requests.
Osirix Web server
Understanding a JPEG 2000 Teaching Server Network
As I alluded above, transmission of data over the network is facilitated by the use of data compression with the JPEG 2000 algorithm. This should not be conflated with the production of jpg' alt='format images. It is a compression scheme that permits lossless and lossy compression. The degree of compression may be stipulated in OsiriX Preferences:General.
The default values may be changed to achieve greater compression, without a perceptual difference in quality. I highly recommend you do this as it will allow you to store more images on your storage medium and your cases will be retrieved faster over the network. Review the guidance document on this web site for details. It's easy!
Here is the default compression scheme I suggest you use:
Just get in the habit of right-clicking on a case after you've anonymized it and choosing 'Compress DICOM files.'
For best performance and transmission efficiency, the network should be configured such that all aspects involve the JPEG 2000 transfer syntax. The intended result is that decompression of the images for viewing will occur once only: after the data have been received by the requesting Workstation. When using the WADO (Web Access for Dicom Objects) protocol for retrieving cases from a server, one should specify, as indicated in the graphics above, transmission via the Original Syntax (Send Transfer Syntax) which, in turn, should be specified (in Preferences: Locations) as JPEG 2000.
For further details about this, read the relevant part of this PDF document
Here is a graphic summary of the requisite setup:
If you're unsure whether a particular case on your server is compressed, compress it manually as shown here:
Querying a Server for Teaching Cases
There are two basic ways of querying a Server for cases and retrieving them:
- Using the OsiriX Web Portal
- Using the OsiriX Query/Retrieval window
The Web Portal
After you login to the Web Portal and find a case you want to retrieve, there are three choices as shown in this graphic:
- WADO retrieval directly to your OsiriX database
- Download case as a Zip file
- Download the case and view it with the Weasis viewer.
WADO retrieval is a great option. Indeed, you could right-click on the OsiriX icon and choose Copy Link Location and send the link to any one with access to your server to allow him to use the link to download that case:
Allowing others to retrieve cases via the Web Portal is a good option because you can control which people have access (username and password) to it and control whether they can upload cases to it or share cases with others. All these options may be specified when you select this method of control:
If you select to view the case with Weasis, the java-based Weasis Viewer will automatically be downloaded to your computer, started, and the case retrieved into it (You have to permit java applications to be downloaded and run on your computer.) . It's also a great way for someone without OsiriX to view the case.
Using the OsiriX Query/Retrieval window
This is a great way to query multiple OsiriX Servers simultaneously as shown here where I'm querying my and a colleague's servers for an 'aortic…' case:
Reminder: Before retrieving a case using this method, check again to be sure you have specified the WADO protocol and Original Syntax in your OsiriX Preferences: Locations for the server you're about to retrieve from.
DICOM-sending a case to a colleague's server
You can DICOM-send a case to another member's server when saving it to our own. This is easy to do. Just select the case in your Database Window and drag-and-drop it over the icon for it in the Sources List as shown here:
Exporting a case to share with colleagues
In addition to the methods for directly sharing cases via the network I described above, you can export a particular anonymized case to share with colleagues, for example, via Dropbox or a similar cloud-based service:
- Create and label a new folder on your desktop.
- Right-click on a case (or select several cases) and choose
Export to DICOM File(s) - Navigate to and select the folder you created on your Desktop and specify the options shown here:
Important: Note the checkbox for Compressing files with JPEG above. Select it. If you've forgotten to compress your cases (as I mentioned above), this will achieve compression at this point.
Now drop the folder into your Dropbox and email them a link to download it once it has been uploaded to the Dropbox server.
[ChimeraX DICOM index]
DICOM Viewers
- Viewers
← Viewers
See also: I Do Imaginglist of free medical imaging software, especiallyMac DICOM display- Medical Holodeck
- DICOM viewer for VR (Vive, Oculus Rift, Windows mixed reality HMDs:Samsung Odyssey, Dell Visor, Lenovo Explorer, Asus Windows, Acer Windows)
- versions: free, basic ($75/mo), pro ($250/mo), cloud ($290/mo);paid versions have 30-day free trial
- Horos
- free and open source (LGPL-3.0) but no free documentation; Google group forum not that extensive
- macOS 10.11+ only, not Windows or Linux
- more...
- for some reason, I cannot download from Firefox (dmg stays at size 0 forever), but download from Safari is fine
- at least to novice, Horos and Osirix Lite interfaces seem very similar(organization, layout, options and corresponding icons)
- File... Import allows selecting a stack of DICOM files or a folder from which multiple stacks found in subdirectories can be opened
- datsets are saved in a Horos database as either file copies or links, then automatically reopened upon next use unless explicitly deleted from that database
- to get to volume visualization from the initial database window:
- click 2D Viewer icon top middle of database window
- from the resulting 2D viewer, choose menu item 3D Viewer... 3D Volume Rendering (other choices include MPR options, 3D MIP, 3D Surface Rendering)
- top bar now allows choosing CLUT or 3D Preset, level of detail, shading, parallel (orthographic) vs. perspective, filtering, etc.:
- a 3D preset (seeplist files in repository)includes not just a CLUT but other settings likeprojection mode, background color, shading, and filtering... perhaps moreeasily understood by clicking Info on the 3D preset browser in Horos
- show histogram w/ thresholds by double-clicking certain entries in the 3D preset browser (I guess the ones with xy pairs in the plist file, e.g. Airways II, Gold Bone, Soft + Skin, rather than just a reference to a separately defined CLUT, e.g. Glossy, Mid Contrast, Red on White)
- CLUT plist files also in repository
- current state can be saved as a 3D preset
- mouse modes:
- defaults: middle mouse translates, right mouse zooms, (left) click-drag adjusts windowing (vertical WL, horizontal WW, diagonal both) in 2D viewer, XY-rotates in 3D viewer; scrolling goes through planes in 2D viewer (Mac trackpad click-drag +cmd does translation, +alt windowing, +shift zoom, +ctrl windowing in 2D, Z-rotation in 3D)
- * values for WL, WW, zoom, etc. reported continuously in text (and color key labels, if color key shown) on window
- in 2D viewer, cursor position is also reported continuously
- 3D volume-viewer icons for changing left-mouse function:
...from left to right, based on experimentation: windowing (WW/WL), XY-translation, zoom, Z-rotation, XY-rotation, rotate around cursor position (? not good because I haven't figured out howto translate in Z), measure distance, add marker, draw circle (maybe it selects contents for later actions but haven't figured out what), hand-draw arbitrary closed shape (again presumably to do something else later), bone removal based on values specified in secondary dialog - 3D surface-viewer mouse modes are only zoom, translate, rotate, center of rotation, and mark; threshold level, resolution (step), and smoothing are set in a secondary dialog, and with smoothing it may not be fast enough for a thresholding mouse mode
- * orientation cube labeled A(nterior), P(osterior), L(eft), R(ight), S(uperior), I(nferior)
- * the cropping box has a little ball in the center of each side for grabbing/dragging
- show color key in 2D viewer with 2D Viewer... Color Look Up Table Bar
- Horos plugins include fairly simple map operations, calculations, and converting other image formats to DICOM
- single-DICOM-file segmentations seem to be ignored (subset of RIDER Lung data from TCIA [zip file]), if I import a directory with both segmentation(s) and a corresponding image stack, I only see the image stack in the database, and even if I explicitly import a single segmentation file, nothing seems to happen.
1/22/2019 I can create a segmentation using the 2D viewer menu ROI... Grow Region (2D/3D Segmentation) and save it as a .roi (2D) or .rois_series (3D) file (poorly made example that goes with the lung dataset) that can be read back into Horos. If it's a 3D segmentation, then ROI... Compute Volume can be used. It thinks a while and then shows a results panel with options including opacity and 'Save as DICOM' but unfortunately that only refers to a single 2D image. Saving lists the image in the database window from where it can be exported to a DICOM file, but again, it's just one 2D image. Can mask data with the 3D segmentation (ROI... Set Pixel Values to). Youtube video not in English but shows some of this stuff.
- my example time series (subset of mouse brain astrocytoma MRI from TCIA [zip file]) is just intepreted as a really big stack, but maybe some other steps are needed to treat/mark as 4D. Clicking the '4D Viewer' icon says that multiple series taken at different times are needed as input (whereas my example series that is 600 .dcm image files all in one directory seems to be misinterpreted as one big series with multiple mouse heads stacked together). (In Horos, had the same problem with the time series example linked to the Slicer MultiVolume Explorer documentation, see below)
- 1/29/2019 Horos displays RTDOSE, RTPLAN (as in the Head-Neck-Cetuximab study from TCIA) as black-and-white stripes in preview/thumbnail, and trying show them in the 2D viewer gives 'no files available (readable) in this series'... although thumbnail images are shown for the CT and RTSTRUCT 'from rtog conversion,' trying to show them in the 2D viewer gives the same complaint
- OsiriX
- OsiriX MD commercial, OsiriX HD for iOS, OsiriX Lite free version
- recent macOS, supports 64-bit computing and multithreading
- basic tutorials available for free, advanced for $$
- 3D Slicer (see alsothe wikiand forum)
- open source 'BSD-style' license, NIH-funded
- 3D Slicer as an image computing platform for the Quantitative Imaging Network.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R.Magn Reson Imaging. 2012 Nov;30(9):1323-41.
- more...
- various filtering options
- interactive and automated segmentation tools
- rigid and non-rigid registration (superposition)
- surface-model creation and manipulation
- users can save scenes (something like sessions?), each potentially containing multiple scene views
- native formats: Slicer scene file is MRML (medical reality modeling language), saved with data as a MRB (medical reality bundle) file. From using the program, looks like the primary option for saving individual image series and segmentations is NRRD (nearly raw raster data). Several other formats also read, tiff, jpeg, vtk, ...
- I tried Slicer 4.10.0 on Mac (desktop and laptop)
- startup quite slow, on desktop always asks if I want to install Xcode
- many tutorials (the PDF slideshows seem nice, the videos vary), including:
- 3D Visualization of DICOM Data
- to display DICOM there are two steps: import the data, then load it from the Slicer DICOM database
- Volume Rendering module has several presets
- the volume rendering becomes low-resolution during manipulation
- in this interface, ROI means a cropping box; the little squares are 'handles' to drag the box faces or translate the box in any 2D view or the 3D view
- if you miss the little square in the 2D view, dragging will instead change windowing: WL (vertical) and/or WW (horizontal)
- then turning volume rendering on by clicking the eye icon only does it within the smaller ROI
- also viewed Quick Start Guide (mainly justhow to install), Welcome, 4-Minute Tutorial for first-time users(looks like more than 4 minutes and mostly showing pre-made VTK surface models,but interesting)
- 3D Visualization of DICOM Data
- module documentation includes:
- Segmentations'It is important to remember that segmentations are not labelmaps'
- Segment Editor – apparently meant to replace Editor(opening Editor shows message to use Segment Editor for more advanced editing)
- Editor – manual segmentation; labelmaps are related to,but not (as mentioned above) the same thing as segmentations
- single-file segmentation DICOM (examples in the RIDER Lung data from TCIA [zip file]) is recognized on import, Modality: SEG in the Slicer DICOM browser; a dialog appeared to suggest I get the Quantitative Reporting module...
- Extension Manager can be opened by clicking the 'E' icon on Slicer window top right.
- QuantitativeReporting depends on other extensions: PETDICOMExtension, DCMQI, SlicerDevelopmentToolbox, so I clicked to install them all.
I tried making my own segmentation in Slicer, as in this video tutorial. There are lots of options, including those analogous to Volume Eraser and Hide Dust. Segmentation save format is only NRRD, and export format choices are only STL and OBJ. Saved NRRD file could be read back in, and was automatically listed as a segmentation. Exported STL file (coordinate system choices LPS or RAS, I chose the default LPS) could be read back in, but was listed as a separate model, not a segmentation, and was not aligned.
1/19/2019 In the RIDER Lung segmentation DICOM file metadata, manufacturer and software version refer only to Slicer, but the manufacturer model field gives the URL for the Quantitative Reporting module, which sounds like it can save DICOM segmentations. I couldn't use it to save the segmentation opened from previously saved NRRD (error: empty segmentation), but I could save one made in the current session or one previously read from DICOM (RIDER Lung). However, 'saving' in this context only means adding it to the internal Slicer DICOM database (SQL), where the RIDER lung segmentation was already. Since the database contents persist through quitting/restarting Slicer, this process at least allows saving my segmentation for later use in Slicer, but doesn't produce a file that can be used in some other program. The Slicer database dialog has an Export button for writing DICOM, but its balloon help says 'not yet available' (even in daily build 1/18/2019) and trying to use it anyway gives 'Error occurred in exporter' message.
I see another video tutorial showing a different set of steps for saving segmentations + volume to DICOM. I followed that entire workflow which worked up to clicking the Export button, which did absolutely nothing, as far as I could tell. However, in the video looks like I need a DicomRtImportExport plugin or something like that. I didn't find exactly that plugin name, but I installed one named SlicerRT (and TCIABrowser too since it interested me). Restarted, repeated the entire process, still got error or nothing exported.
1/20/2019 Got a little farther: if I choose export format RT, I can export a study 'to DICOM' using the workflow shown in that video, but only to the Slicer DICOM database and only as modality RTSTRUCT, not SEG. Export type DICOMSegmentation seems to either cause error or do nothing, export type Scalar Volume only exports the images, not the segmentation. Again, no actual DICOM output files, only export to the Slicer DICOM database. The only way I found to get SEG into the database is with the Quantitative Reporting module, as reported above.
- time series (subset of mouse brain astrocytoma MRI from TCIA [zip file]) was read without error messages; need to investigate further.
1/22/2019 Now I see it is loaded as a 50-frame 'multivolume,' apparently their term for a time series, in agreement with the 50-frame time slider in ChimeraX, and can be played using the MultiVolume Explorer module. Another example time series (prostate) is provided in that module's documentation.
- 1/29/2019 Slicer displays RTDOSE as colored blobs if associated RTPLAN is also opened, RTSTRUCT as colored overlay if associated CT is also opened
2/12/2019 Slicer can render RTSTRUCT in 3D as lines or multiple transparent surfaces, in addition to the overlays on the 2D slice views. Settings/options for creating surfaces from contours are described in the Segment Editor documentation.
- MRIcro family of programs
- open source BSD license, NIH-funded
- MRIcroGL real-time interactive volume rendering, requires modern graphics card (Windows, Mac, Linux)
- these Mac downloads each include two versions:
- [Nov 2018 github] ...includes MRIcroGL and MRIcroMTL (uses Apple's Metal instead of OpenGL)
- [Jun 2018 NITRC] ...includes MRIcroGL and MRIcro for OSX which I guess has lower graphics demands (for macOS 10.7+ 64-bit Intel), manual for MRIcroGL (PDF, Jun 2017), and several sample datasets and templates for overlays
- MRIcroGL is different in those two downloads: color-scheme menu in a different place, the github one has fewer import options and shader options, etc.
- GLSL volume ray casting rendering
- ...* surface renderings do look really nice
- formats: primarilyNIfTI (Neuroimaging Informatics Technology Initiative: .nii, .nii.gz, .hdr/.img).Release notes also mention importing other formats:Freesurfer images (MGH/MGZ format), NRRD images (NRRD/NHDR format), AFNI (HEAD format), VTK/ITK MetaIO format (MHA/MHD), TIFF/LSM/OME.
- does not read DICOM directly, comes with slightly unfriendly (command-line) DICOM → NIfTI converter ...apparently this conversion is quite complex, the author even wrote a paper about it in 2016
- video tutorials page links broken but a few are available at Chris Rorden's YouTube channel
- full colormap control for rendering different tissues differently
- shaders are text files, users can create their own custom combinations
- Pascal scripting
- I tried MRIcroGL on 2015 MacBook Pro and 2012 iMac running High Sierra (10.13): nice graphics, smooth response to manipulation, LOTS of rendering options, sliders for individual parameters, many built-in color schemes (some with intensity values for specific tissues in CT), clipping, cutouts (box-shaped), slice views, overlays ...
- with both program versions, trying to change color scheme for the 'visible human' dataset didn't change its appearance, but other example datasets didn't have this issue... maybe because that dataset consists of thin-section (color?) photographs rather than monochrome imaging
- automatically remembers/opens last data, opening something else replaces it
- as far as I can tell, one program instance shows one dataset (potentially with overlay(s)), but multiple instances can be run simultaneously and 'yoked' to synchronize slice display
- click-drag rotates, orientation cube shown, scrolling zooms (up to a limit); no panning?
- use keys a(nterior), p(osterior), l(eft), r(ight), s(uperior), i(nferior) to move slice plane
- prefer Chimera(X) interactive clip translation/rotation over the 3 sliders
- * useful predefined color scheme values for specific tissues (see below)
- * nice color key automatically shown/updated (yet optional)
- used the DICOM → NIfTI converter on one of the Belarus chest datasets; got some errors/warnings, some .nii output didn't look right, but the last/largest one appears to have all the data: bones, lungs and other tissues shown nicely with the appropriate color schemes
- tried to import Allen Inst. OME TIFF but got error about incompatible compression type
- update 2 September 2019: latest release supports drag-and-drop to open DICOM folders! Also has expanded TIFF support (now can open my Allen Inst. OME TIFF example) and Python scripting.
- these Mac downloads each include two versions:
- Web-based viewers:
- OHIF (Open Health Imaging Forum)
- many basic Windows-only DICOM viewers that I won't bother to list here
← Acronyms & Terminology
See also: DICOM modality attributes
- ADC – apparent diffusion coefficient
- AIM – Annotation and Image Markupeffort to standardize medical image annotation (NIH/NCI)
- CR – computed radiography
- CT – computed tomography
- CLUT – color lookup table
- DCE – dynamic contrast enhancement
- DICOM – file standard for digital imaging and communications in medicine
- DTI – diffusion tensor [magnetic resonance] imaging
- DWI – diffusion-weighted [magnetic resonance] imaging
- Hounsfield scale – normalized units used in medical CT
- LPS – left, posterior, superior anatomical coordinates
- RAS – right, anterior, superior anatomical coordinates
- MIP – maximum intensity projection
- MPR – multiplanar reconstruction or reformatting (generating 'slices' in arbitrary directions or even curved)
- MRI – magnetic resonance imaging
- PACS – picture archive and communication system
- PET or PT – positron emission tomography
- QIN – Quantitative Imaging Network (NIH/NCI)
- ROI – region of interest
- RT – radiotherapy
- structure set – set of areas of significance
- SR – structured report
- SUV – standardized uptake values (PET/CT)
- WL – window level
- WW – window width
← Feature Lists
Potential tasks for medical image analysis:
Osirix Free Download For Mac
- view header information
- anonymize
- zoom, rotate, flip
- invert dark/light
- adjust contrast and brightness
- color processing, color mask
- adjust windowing/thresholding #1613
12/17/18 Darrell mentions 'color lookup table (CLUT) editor,' HOROS has mouse modes to adjust:- leveling - moving thresholds in parallel along histogram
- windowing - narrowing/broadening visible range of data values (moving bounding thresholds in opposite directions)
(Elaine: maybe labeled color key as inMRIcroGL or Horos,WW/WL values as in Horos)
(related: Phil mentions popup reporting data value at voxel like HOROS) - built-in color schemes:
- HOROS 3D presets (he often uses Airways II)
- convert data types, formats
- filter (smooth, sharpen, etc.)
- other data editing (copy, paste, erase?), Chimera Volume Eraser
- clipping
- mark points and regions
- select regions, potentially of irregular shape
- detect/mark edges, boundary surfaces; mask and segment
12/17/18 Beth and Darrell mention segmentation, particularly by hand marking - annotate (text or geometric objects in image, metadata outside of image)
- geometric measurements (distance, angle, area, volume, etc.)
- data measurements (mean, SD, min, max, plot values along an axis, etc.)
- texture analysis (not sure what this means)
- play time series, save movie files
- compare datasets (with and without contrast agent enhancement;before, during, and after therapy; normal vs. abnormal brain; etc.)
- overlays (annotations, localized brain activity, etc.)
- registration with atlases/templates (anatomical landmarks, referenceorgans) or between multiple datasets, rigid or non-rigid (with scaling/warping)
- along those lines, medical image fusion...essentially 3D superposition of multiple datasets or maps
- fetch data from public sources
Other desirables from discussion:
- large data performance, especially in VR
- multi-user VR meeting
- working with multichannel data, e.g. annotations from AI/deeplearning may be saved as color channels to be overlaid on the exptl. dataset
Osirix Free Download
← Public Sources of Files and Info
- The Cancer Imaging Archive (TCIA) has many collections ofdatasets (public collections also listed onthe wiki)
- formerly the National Biomedical Imaging Archive (NBIA)
- downloading requires NBIA Data Retriever app
- however, there's also aREST APIfor programmatic access
- Patient Contributed Image Repository(only 10 downloads)
- MedicalImage Samples (only 5 multiframe sets)
- open-access medical image repositories – a big list,but images are not necessarily in DICOM format
- NIH DeepLesion dataset – over 32,000 annotated lesionsin CT images available via Box, but currently only PNG format
Osirix Mac
- David Clunie's Medical Image Format Site(some files in Images section near bottom)
